Genetic and environmental markers of cancer therapy
Leverage high-dimensional clinical and biological data to decipher resistance to cancer immunotherapy
Immunotherapy is one of the major treatments for patients diagnosed with cancer. However, on average less than half of the patients showed clinical benefit. Known biomarkers of response include a T cell-inflamed tumor phenotype characterized by CD8+ T cells and type I/II interferon expression, and a high tumor mutational burden (TMB). Our primary focus is to identify tumor-intrinsic oncogenic pathways that drive immune exclusion in tumor microenvironment (TME) and lack of response to immune checkpoint inhibitors (ICI). Our previous work established an atlas of somatic mutations and activated pathways that contribute to a non-T cell-inflamed phenotype across human solid tumors. We demonstrated an increasing number of co-activated pathways leads to more highly non-T cell-inflamed tumors. To further pursue this question, we extended our work to internally annotated patient cohorts diagnosed with melanoma, NSCLC, RCC, and other cancer types and treated with anti-PD1 with rich clinical data. We perform high-throughput bulk (RNAseq, WES) and single-cell sequencing (10X), coupled with high-dimensional imaging and spatial transcriptomics/proteomics profiling from human specimens, with the ultimate goal to interrogate mechanisms underlying tumor cell-derived modulation of T cell and other immune cell subset infiltration in TME. Our work provides a personalized multi-omics approach to detect mechanisms that may contribute to resistance to cancer immunotherapy. We have translated our findings to new immune-potentiating interventions, including a novel combination clinical trial that we recently launched for patients with IDH1 mutant tumors (see section below “Innovative Clinical Trials”).
Determine microbial and metabolite modulates of tumor-immune microenvironment and anti-PD1 efficacy
A high level of tumor-infiltrating CD8+ T cells is a known biomarker for response to ICI. Our group, among others, discovered that specific gut microbes modulate T cell response and anti-PD1 efficacy in metastatic melanoma. Emerging evidence supports a high prevalence of tumor type-specific microbiota residing within tumor tissues, and these are disproportionally enriched in melanoma of responders and non-responders to ICI. We hypothesize that microbiota from the gut or tumor may cross-talk with each other and modulate the anti-tumor immune response in TME. Specific species from the gut and tumor microbiomes are associated with a higher level of tumor-infiltrating CD8+ T cells, tumor-immune cell interactions, and improved response to ICI. To pursue this question, we collect fecal samples and tumor biopsies (melanoma, HNSCC, NSCLC, and others) from patients treated with anti-PD1, and perform microbiome sequencing (16S and shotgun), metabolite profiling (LC-MS/MS and GC-MS), and spatial imaging / profiling on tumor tissue slides to interrogate the interactions between species, microbe-derived metabolites, and tumor/immune cells in contribution to shaping the TME. Our findings will enhance the understanding of the role of microbiomes in cancer immunotherapy.
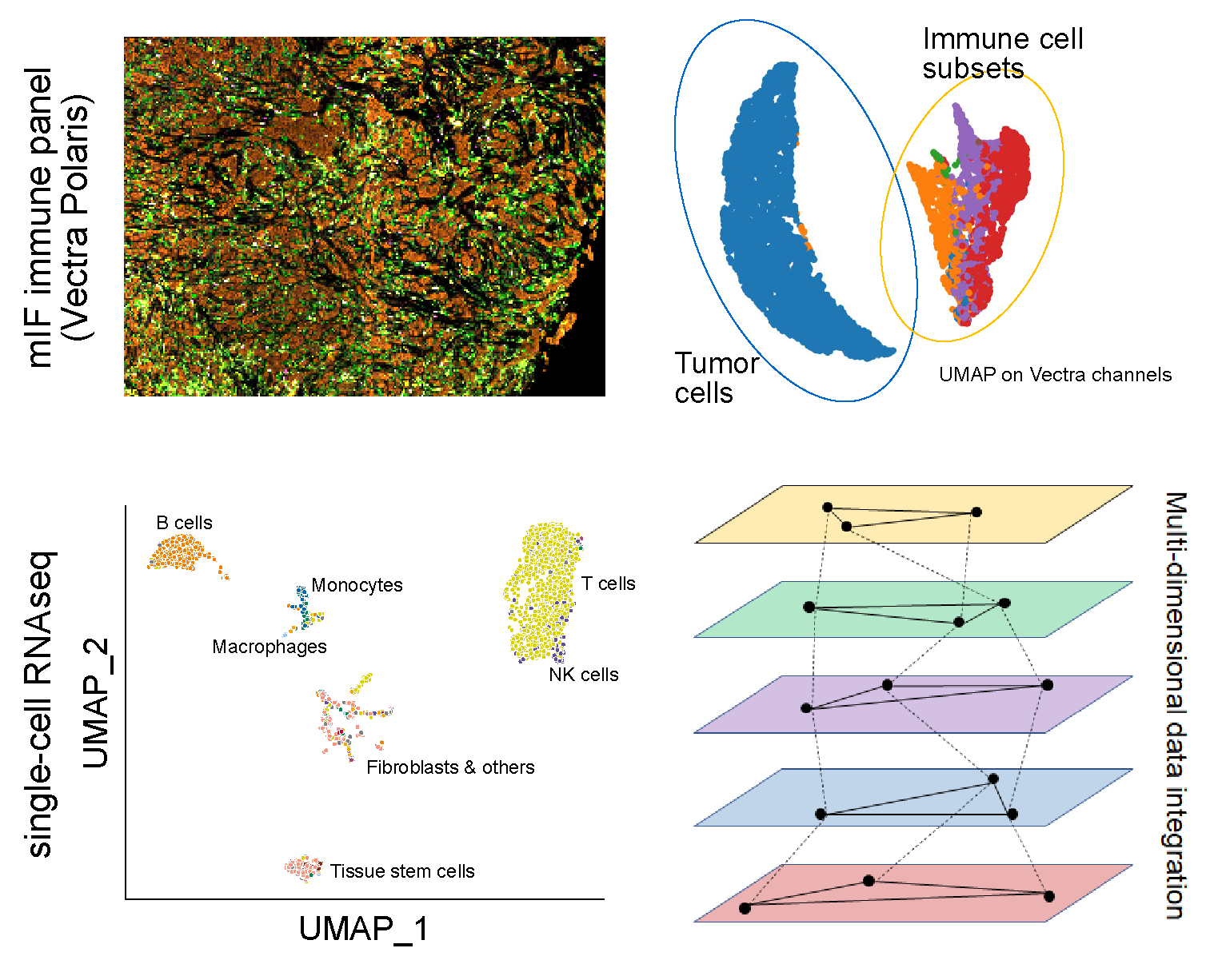
Genomics and spatial imaging of immunotherapy
Computer vision-assisted image pattern recognition
While sequencing has been extensively employed as a technology for biomarker discovery, recent development in multiplex imaging techniques and GPU-accelerated computing algorithms have enabled the discovery of spatial biomarkers from direct capture of TME snapshots on tumor tissues slides. We are strongly invested in technology advances in imaging processing and recognition, including: (1) Develop deep-learning and machine-learning models to extract features from high-resolution (20X or 40X) H&E, DSP, and mIF images (Vectra, CODEX), (2) Design and implement new segmentation approaches to distinguish overlapping cells and regions of high cell density with internally annotated gold-standard datasets and build multiple-channel models for better marker+/- cell subset classification, (3) Establish co-registration techniques to align signals across sectioned slides, enabling comparison of marker+/- immune and tumor cell subsets across panels.
Interrogate tumor-immune cell-cell spatial interactions in contribution to resistance mechanisms accounting for tumor heterogeneity
Tumor heterogeneity is a huge barrier to developing accurate and biologically interpretable models to understand resistance to ICI. To address this challenge, We use state-of-art technologies to interrogate the interactions among malignant, immune, and stromal cells in TME at single-cell resolution. We perform spatial imaging (Nanostring GeoMX Digital Spatial Profiling, Vectra, CODEX) and single-cell sequencing (10X scRNAseq, scTCRseq, etc.) on human specimens to identify tumor cell subclones, tumor-immune cell-cell distance, and tumor-immune tertiary structure contributing to a non-T cell-inflamed TME and lack of response. We then integrate the spatial elements from imaging & single-cell experiments with genomic markers and clinical annotation to identify new resistance mechanisms accounting for tumor heterogeneity, and develop improved classification models to predict response to ICI.
Innovative clinical trials
Multi-Center Phase I/II Trial of Encorafenib with and without Binimetinib in Combination with Nivolumab and Low-dose Ipilimumab in Metastatic BRAF-mutant Melanoma
Patients with high risk BRAF mutant metastatic melanoma, such as those with brain or liver metastases, elevated lactate dehydrogenase (LDH) and bulky disease, have inferior treatment outcomes with current therapies. This is an open label, multi-site, Phase 1/2 study of BRAF +/- MEK inhibition + anti-PD1 + low dose anti-CTLA4 for BRAF-mutated melanoma in high-risk cohorts (CT.gov: NCT04655157). Continuous Bayesian toxicity monitoring will be used throughout to monitor DLT. Pre and on-treatment tumor biopsies (WES, RNAseq, scRNAseq) will assess changes in the tumor microenvironment while peripheral blood ctDNA and T cell Ki67% changes will assess early response and immune activation during triplet and quadruplet therapy.
Phase I Study Investigating the Safety of Stereotactic Body Radiotherapy (SBRT) with Anti-PD-1 and Anti-IL-8 for the Treatment of Multiple Metastases in Advanced Solid Tumors
Our previous studies combining multi-site SBRT with immuno-oncology (IO) agents have revealed induction of IFN signaling by SBRT and serum IL8 (sIL8) as strongly associated with lack of response to RT-IO. Overcoming IL8 induced epithelial-mesenchymal transitioning and trafficking of myeloid derived suppressor cells in the TME represents a promising strategy to overcome resistance. BMS-986253 (anti-IL8) with nivolumab has demonstrated safety and preliminary activity. We are investigating BMS-986253 + nivolumab + SBRT (CT.gov: NCT04572451) in solid tumors, RCC and melanoma via Bayesian toxicity monitoring for DLT and efficacy assessment. We have integrated analysis of WES and RNAseq from baseline tissues as well as analysis of serum IL-8 levels and radiation-induced changes in peripheral blood T cell populations with response to treatment.
A Phase Ib/II study of ARRY-614 plus either nivolumab or ipilimumab in advanced melanoma,
RCC, and solid tumors
Inadequate dendritic cell activation and cytokine support has been linked to ineffective immune checkpoint blockade. p38 MAPK has been identified as a suppressor of DC activity and inhibition of p38 may enhance immunotherapy. We are pursuing a safety and signaling finding Phase Ib/II study of p38/TIE2 inhibition in combination with checkpoint blockade in solid tumors, melanoma and RCC (CT.gov: NCT04074967). We have are additionally pursuing genomic (WES, RNAseq) and peripheral blood (ctDNA, immune profiling) studies of treatment response from baseline and on-treatment biospecimens.
Phase II study of IDH1 inhibitor ivosidenib and nivolumab in IDH1 mutant gliomas and advanced
solid tumors
In our prior work, we have identified IDH1 mutation as a pan-cancer mediator of the non-T cell-inflamed phenotype and immune exclusion. We are pursuing a study of genomically defined patients where we will explore the safety, early efficacy as well as immune and metabolomic effects of IDH1 inhibition + anti-PD1 (CT.gov NCT04056910). Will pursue WES, RNAseq, scRNAseq and ChIP-Seq from baseline and on-treatment biopsies. We will further evaluate changes in ctDNA and immune monitoring of circulating populations to describe the impact of this novel treatment approach.